An Essential Toolbox for RawSequencing Data Analysis
Sarani Santerne
As you might have noticed, bioinformatics is a vast field. However, there is one common analysis which you can encounter quite frequently and can help you get started: the raw sequencing data analysis. As all these steps can be confusing sometimes, I propose a generic walkthrough in order to give you a few basics to start your project.
Summary workflow
As a side note, I advise you to create workflows to document your projects: I find it clear and informative enough. Below is a generic example for today’s insights:
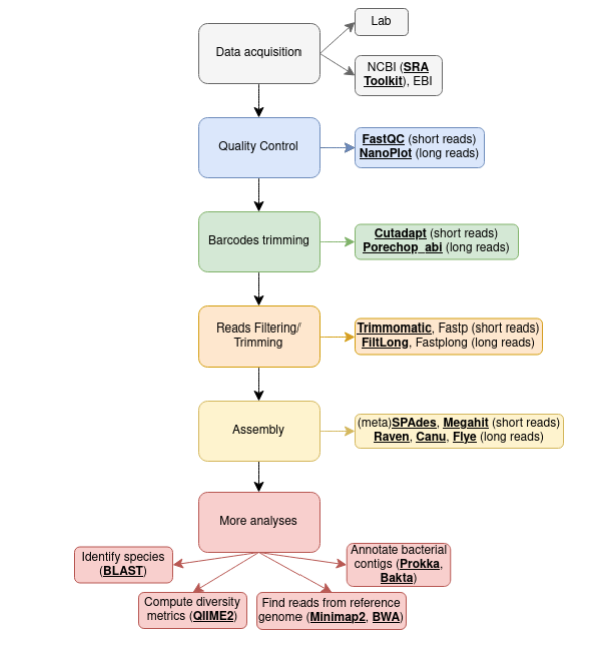
Here, I won’t go into deep details about the use of every single command. My goal is to provide you with tools which you can use in specific situations for the process of analyzing raw sequencing data, and it will be your duty to then check their documentation and see how you can use them in your case.
Data acquisition
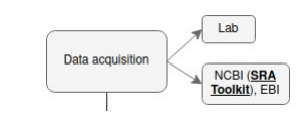
Whether you are working in a company or a lab and will be getting your data straight out of the sequencer, or you want to work solo on a project and will be needing data to play with.
You have a few common ways to find sequencing data online: the best starting point is to narrow down your searches by picking a research paper and find the provided accession number for the raw data. Once you know your organism of interest, you can:

Don’t forget to check the data’s characteristics (paired-end, single-end, NGS, ONT/PacBio…). This information is available on the sequencing report if you got the data from a lab, or in the article/EBI description. It will be used in the analysis, as tools often require a specific option for long reads, or influence your choice of tools directly.
Data Quality Control (QC)
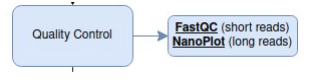
First things first: estimate how good your data is by generating reports containing graphs and metrics before proceeding any further.
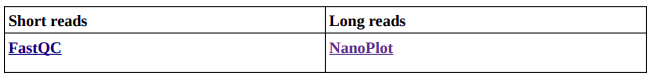
See, retaining information about the sequencing data already comes in handy to choose which tool to use!
This quality check helps determine whether your reads need filtering and trimming. This is at your own discretion and depends on your data’s quality. You can determine it by checking where does the quality lowers on the plots from the tools’ reports.
Barcodes Trimming
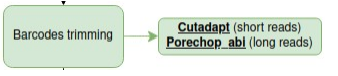
You may want to eliminate barcodes in your samples, as it could become a noise or add extra Mbs of data later on.
Reads Filtering/Trimming
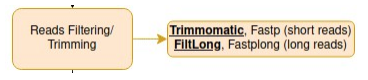
It is time to filter your reads based on a minimal read length and minimal quality determined earlier.

If you work on metagenomics samples, you can deplate the host’s DNA using Bowtie2.
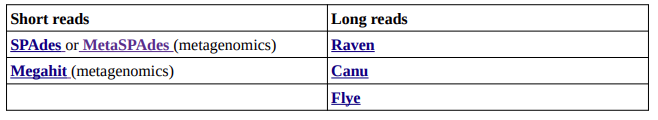
Assembly
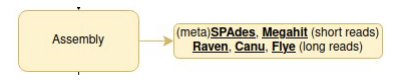
Now that you have your clean data, it’s time to make a puzzle! Right now, you only have sequenced reads, but you might want to identify what organisms are there, and thus assemble these reads into longer contigs. You have different assembly methods, which I won’t detail, but remember that several tools can use different assembly methods, and give different results.
Now what?
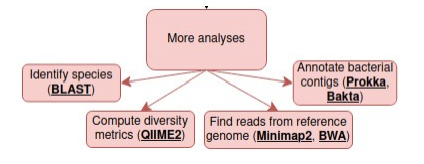
Now that you have your data, it depends on what you want to do with it – here are a few leads to get you started.
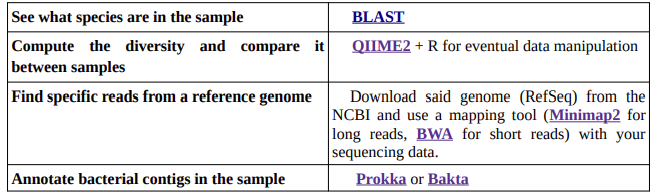
More useful tools
Take Home Message
The main message I would like to convey here is: if you begin in bioinformatics, remember that you don’t have to code every single function, as they probably already exist – and are optimized to be faster. Thus, always verify if you really need to code it or if you can use an already developed tool.
There’s no shame in that!
Also, remember that sometimes, your commands won’t work right away, or you will have doubts regarding what options to use. In this case, always stay curious and do check the documentation and forums before turning to ChatGPT for command generation! Use AI wisely: it can help, but it’s only an LLM basing itself on training data and attention mechanisms. The more you write commands on your own and decide on what tools to use based on your own insights, the more experienced you
will become.
References
For any work you accomplish, always credit their initial authors, whether it is github links and/or papers.
SRA Toolkit Development Team. https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=software
https://github.com/ncbi/sra-tools
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. https://github.com/s-andrews/FastQC
Wouter De Coster, Rosa Rademakers, NanoPack2: population-scale evaluation of long-read sequencing data, Bioinformatics, Volume 39, Issue 5, May 2023, btad311.
https://github.com/wdecoster/NanoPlot
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. https://cutadapt.readthedocs.io/en/stable/
Bonenfant, Q., Noé, L., Touzet, H. (2022). Porechop_ABI: discovering unknown adapters in ONT sequencing reads for downstream trimming. https://github.com/bonsai-team/Porechop_ABI
Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics, btu170. http://www.usadellab.org/cms/?page=trimmomatic
Shifu Chen, Yanqing Zhou, Yaru Chen, Jia Gu; fastp: an ultra-fast all-in-one FASTQ preprocessor, Bioinformatics, Volume 34, Issue 17, 1 September 2018, Pages i884–i890. https://github.com/OpenGene/fastp https://github.com/rrwick/Filtlong
Shifu Chen. 2023. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2: e107. https://github.com/OpenGene/fastplong
Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012, 9:357-359. https://bowtie-bio.sourceforge.net/bowtie2/index.shtml
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A., & Korobeynikov, A. (2020). Using SPAdes de novo assembler. Current Protocols in Bioinformatics, 70, e102. https://github.com/ablab/spades
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. . 27, 824–834 (2017).
Li, D., Liu, C-M., Luo, R., Sadakane, K., and Lam, T-W., (2015) MEGAHIT: An ultra-fast singlenode solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. https://github.com/voutcn/megahit
Vaser, R., Šikić, M. Time- and memory-efficient genome assembly with Raven. Nat Comput Sci 1, 332–336 (2021). https://github.com/lbcb-sci/raven
Koren S, Walenz BP, Berlin K, Miller JR, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Research. (2017).
https://github.com/marbl/canu
Kolmogorov, M., Yuan, J., Lin, Y. et al. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37, 540–546 (2019).
https://github.com/mikolmogorov/Flye/blob/flye/docs/USAGE.md
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410.
https://www.ncbi.nlm.nih.gov/books/NBK279690/
Bolyen E, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology 37:852–857. https://qiime2.org/
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics, 34:3094-3100. https://github.com/lh3/minimap2
Li H. and Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics, 25:1754-60. https://bio-bwa.sourceforge.net/
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014 Jul 15;30(14):2068-9. https://github.com/tseemann/prokka
Schwengers O., Jelonek L., Dieckmann M. A., Beyvers S., Blom J., Goesmann A. (2021). Bakta:rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microbial Genomics, 7(11). https://github.com/oschwengers/bakta
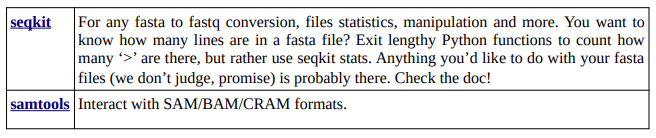
Wei Shen*, Botond Sipos, and Liuyang Zhao. 2024. SeqKit2: A Swiss Army Knife for Sequence and Alignment Processing. iMeta e191. https://bioinf.shenwei.me/seqkit/
Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H, Twelve years of SAMtools and BCFtools, GigaScience (2021). http://www.htslib.org/
Biotational – The Open-Access Hub for Computational Science
Collaborate. Share. Innovate.
Website: https://www.biotational.com
Email: info@biotational.com
LinkedIn: https://www.linkedin.com/company/biotational/
© 2025 Biotational. All Rights Reserved.
This article is published under a Creative Commons CC BY-NC license, allowing for non-commercial sharing with proper attribution.
Want to share your research? Submit your article on Biotational today by emailing info@biotational.com!
Author Contacts: Sarani Santerne | LinkedIn